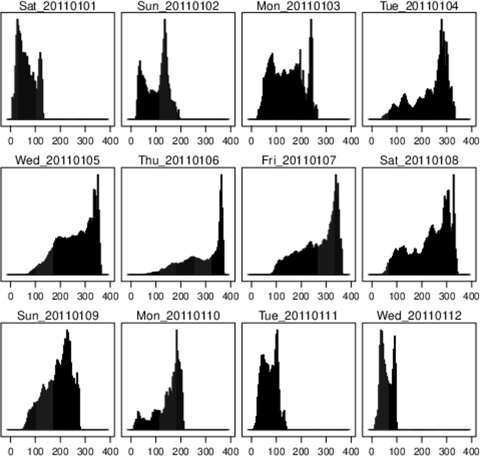Spatio-temporal data - Displaying time series, spatial and space-time data with R
Table of Contents
3. Spatiotemporal Raster data
5. Code
5.1. Spatiotemporal point data
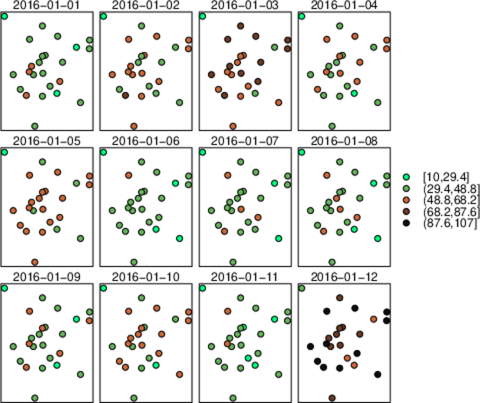
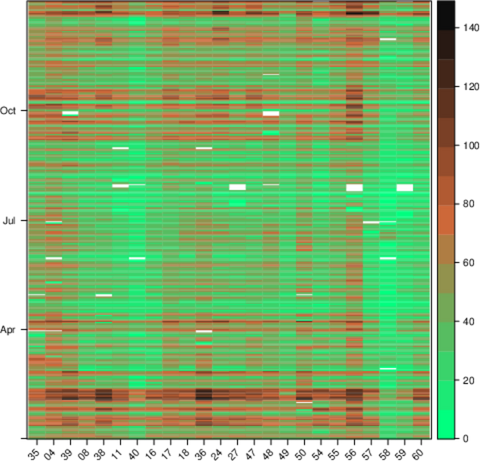
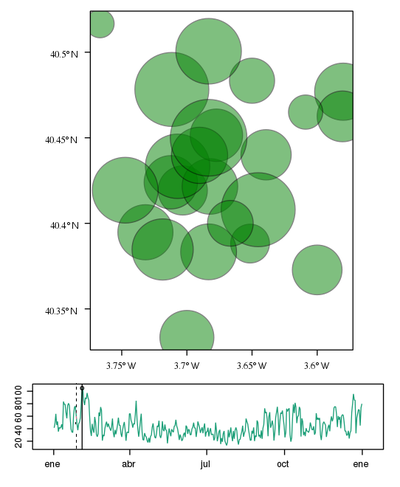
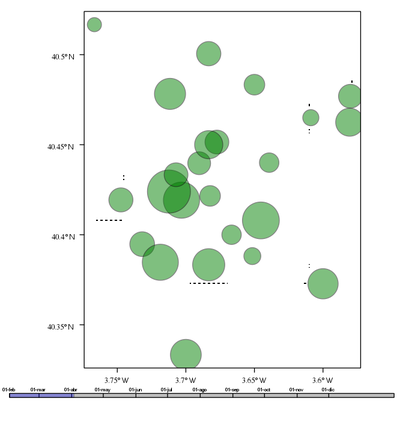
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library("lattice") library("ggplot2") ## latticeExtra must be loaded after ggplot2 to prevent masking of its ## `layer` function. library("latticeExtra") library("RColorBrewer") source('configLattice.R') Sys.setlocale("LC_TIME", "C") ################################################################## ## Data and spatial information ################################################################## ## Spatial location of stations airStations <- read.csv2("data/Spatial/airStations.csv") ## Measurements data airQuality <- read.csv2("data/Spatial/airQuality.csv") ## Only interested in NO2 NO2 <- airQuality[airQuality$codParam == 8, ] library("zoo") library("reshape2") library("sp") library("spacetime") library("sf") library("sftime") ################################################################## ## sp and spacetime ################################################################## airStationsSP <- airStations ## rownames are used as the ID of the Spatial object rownames(airStationsSP) <- substring(airStationsSP$Code, 7) coordinates(airStationsSP) <- ~ long + lat proj4string(airStationsSP) <- CRS("+proj=longlat +ellps=WGS84") NO2$time <- as.Date(with(NO2, ISOdate(year, month, day))) NO2wide <- dcast(NO2[, c('codEst', 'dat', 'time')], time ~ codEst, value.var = "dat") NO2zoo <- zoo(NO2wide[,-1], NO2wide$time) dats <- data.frame(vals = as.vector(t(NO2zoo))) NO2st <- STFDF(sp = airStationsSP, time = index(NO2zoo), data = dats) ################################################################## ## sf and sftime ################################################################## airStationsSF <- st_as_sf(airStations, coords = c("long", "lat"), crs = 4326) NO2$time <- as.Date(with(NO2, ISOdate(year, month, day))) idx <- match(NO2$codEst, airStationsSF$Code) NO2sft <- st_sftime(dat = NO2$dat, code = NO2$codEst, geometry = airStationsSF$geometry[idx], time = NO2$time) ################################################################## ## Graphics with spacetime and sftime ################################################################## airPal <- colorRampPalette(c("springgreen1", "sienna3", "gray5"))(5) stplot(NO2st[, 1:12], cuts = 5, col.regions = airPal, main = "", edge.col = "black") airPal <- colorRampPalette(c("springgreen1", "sienna3", "gray5"))(5) ggplot(NO2sft) + geom_sf(aes(color = dat)) + facet_wrap(~ cut(time, 12)) + scale_colour_stepsn(n.breaks = 6, colours = airPal) stplot(NO2st, mode = "xt", col.regions = colorRampPalette(airPal)(15), scales = list(x = list(rot = 45)), ylab = "", xlab = "", main = "") ggplot(NO2sft) + geom_tile(aes(x = as.factor(code), y = time, fill = dat)) + scale_fill_stepsn(colours = airPal, n.breaks = 6) + xlab("Station Code") + ylab("") + guides(x = guide_axis(angle = 45)) stplot(NO2st, mode = "ts", xlab = "", lwd = 0.1, col = "black", alpha = 0.6, auto.key = FALSE) ggplot(NO2sft) + geom_line(aes(x=time, y = dat), colour = "black", linewidth = 0.25, alpha = 0.6) + theme_bw() + xlab("") ################################################################## ## Point space-time data ################################################################## ################################################################## ## Initial snapshot ################################################################## library("sp") library("zoo") library("reshape2") library("spacetime") airStationsSP <- read.csv2("data/Spatial/airStations.csv") rownames(airStationsSP) <- substring(airStationsSP$Code, 7) coordinates(airStationsSP) <- ~ long + lat proj4string(airStationsSP) <- CRS("+proj=longlat +ellps=WGS84") airQuality <- read.csv2("data/Spatial/airQuality.csv") NO2 <- airQuality[airQuality$codParam == 8, ] NO2$time <- as.Date(with(NO2, ISOdate(year, month, day))) NO2wide <- dcast(NO2[, c("codEst", "dat", "time")], time ~ codEst, value.var = "dat") NO2zoo <- zoo(NO2wide[,-1], NO2wide$time) dats <- data.frame(vals = as.vector(t(NO2zoo))) NO2st <- STFDF(sp = airStationsSP, time = index(NO2zoo), data = dats) library("grid") library("gridSVG") ## Initial parameters start <- NO2st[,1] ## values will be encoded as size of circles, ## so we need to scale them startVals <- start$vals/5000 nStations <- nrow(airStationsSP) days <- index(NO2zoo) nDays <- length(days) ## Duration in seconds of the animation duration <- nDays*.3 ## Auxiliary panel function to display circles panel.circlesplot <- function(x, y, cex, col = "gray", name = "stationsCircles", ...) { grid.circle(x, y, r = cex, gp = gpar(fill = col, alpha = 0.5), default.units = "native", name = name) } pStart <- spplot(start, panel = panel.circlesplot, cex = startVals, scales = list(draw = TRUE), auto.key = FALSE) pStart ################################################################## ## Intermediate states to create the animation ################################################################## ## Color to distinguish between weekdays ('green') and weekend ## ('blue') isWeekend <- function(x) {format(x, "%w") %in% c(0, 6)} color <- ifelse(isWeekend(days), "blue", "green") colorAnim <- animValue(rep(color, each = nStations), id = rep(seq_len(nStations), nDays)) ## Intermediate sizes of the circles vals <- NO2st$vals/5000 vals[is.na(vals)] <- 0 radius <- animUnit(unit(vals, "native"), id = rep(seq_len(nStations), nDays)) ## Animation of circles including sizes and colors grid.animate("stationsCircles", duration = duration, r = radius, fill = colorAnim, rep = TRUE) ################################################################## ## Time reference: progress bar ################################################################## ## Progress bar prettyDays <- pretty(days, 12) ## Width of the progress bar pbWidth <- .95 ## Background grid.rect(.5, 0.01, width = pbWidth, height = .01, just = c("center", "bottom"), name = "bgbar", gp = gpar(fill = "gray")) ## Width of the progress bar for each day dayWidth <- pbWidth/nDays ticks <- c(0, cumsum(as.numeric(diff(prettyDays)))*dayWidth) + .025 grid.segments(ticks, .01, ticks, .02) grid.text(format(prettyDays, "%d-%b"), ticks, .03, gp = gpar(cex = .5)) ## Initial display of the progress bar grid.rect(.025, .01, width = 0, height = .01, just = c("left", "bottom"), name = "pbar", gp = gpar(fill = "blue", alpha = ".3")) ## ...and its animation grid.animate("pbar", duration = duration, width = seq(0, pbWidth, length = duration), rep = TRUE) ## Pause animations when mouse is over the progress bar grid.garnish("bgbar", onmouseover = "document.documentElement.pauseAnimations()", onmouseout = "document.documentElement.unpauseAnimations()") grid.export("figs/SpatioTime/NO2pb.svg") ################################################################## ## Time reference: a time series plot ################################################################## library("lattice") library("latticeExtra") ## Time series with average value of the set of stations NO2mean <- zoo(rowMeans(NO2zoo, na.rm = TRUE), index(NO2zoo)) ## Time series plot with position highlighted pTimeSeries <- xyplot(NO2mean, xlab = "", identifier = "timePlot") + layer({ grid.points(0, .5, size = unit(.5, "char"), default.units = "npc", gp = gpar(fill = "gray"), name = "locator") grid.segments(0, 0, 0, 1, name = "vLine") }) print(pStart, position = c(0, .2, 1, 1), more = TRUE) print(pTimeSeries, position = c(.1, 0, .9, .25)) grid.animate("locator", x = unit(as.numeric(index(NO2zoo)), "native"), y = unit(as.numeric(NO2mean), "native"), duration = duration, rep = TRUE) xLine <- unit(index(NO2zoo), "native") grid.animate("vLine", x0 = xLine, x1 = xLine, duration = duration, rep = TRUE) grid.animate("stationsCircles", duration = duration, r = radius, fill = colorAnim, rep = TRUE) ## Pause animations when mouse is over the time series plot grid.garnish("timePlot", grep = TRUE, onmouseover = "document.documentElement.pauseAnimations()", onmouseout = "document.documentElement.unpauseAnimations()") grid.export("figs/SpatioTime/vLine.svg")
5.2. Spatiotemporal areal data
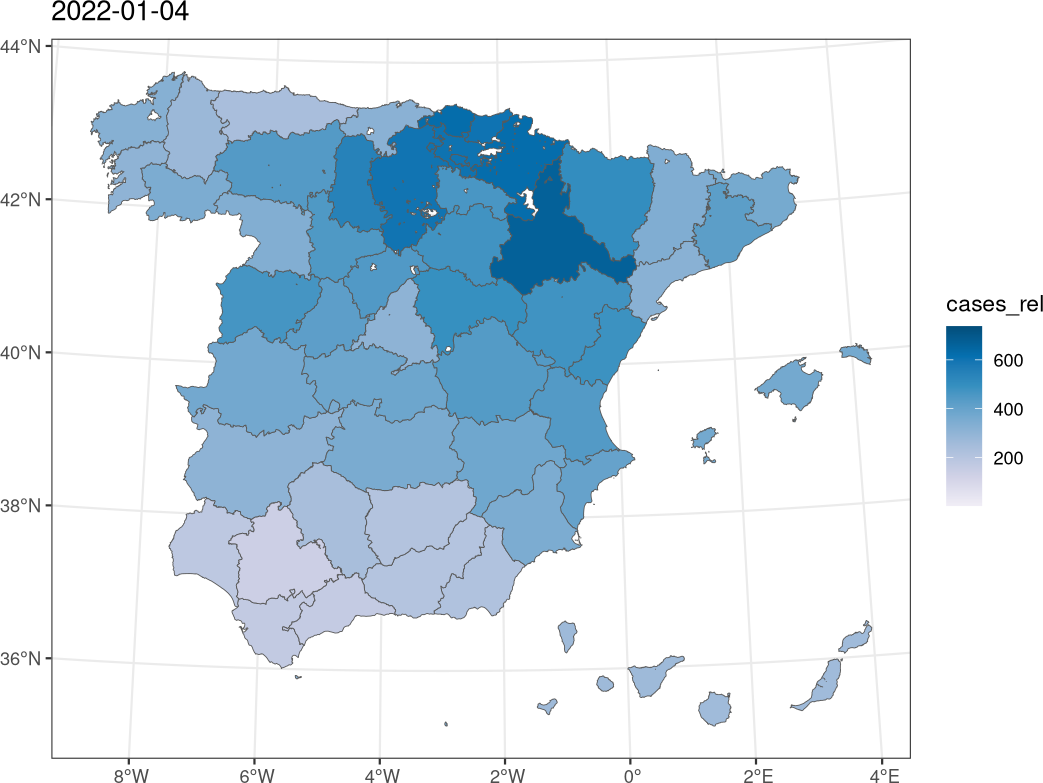
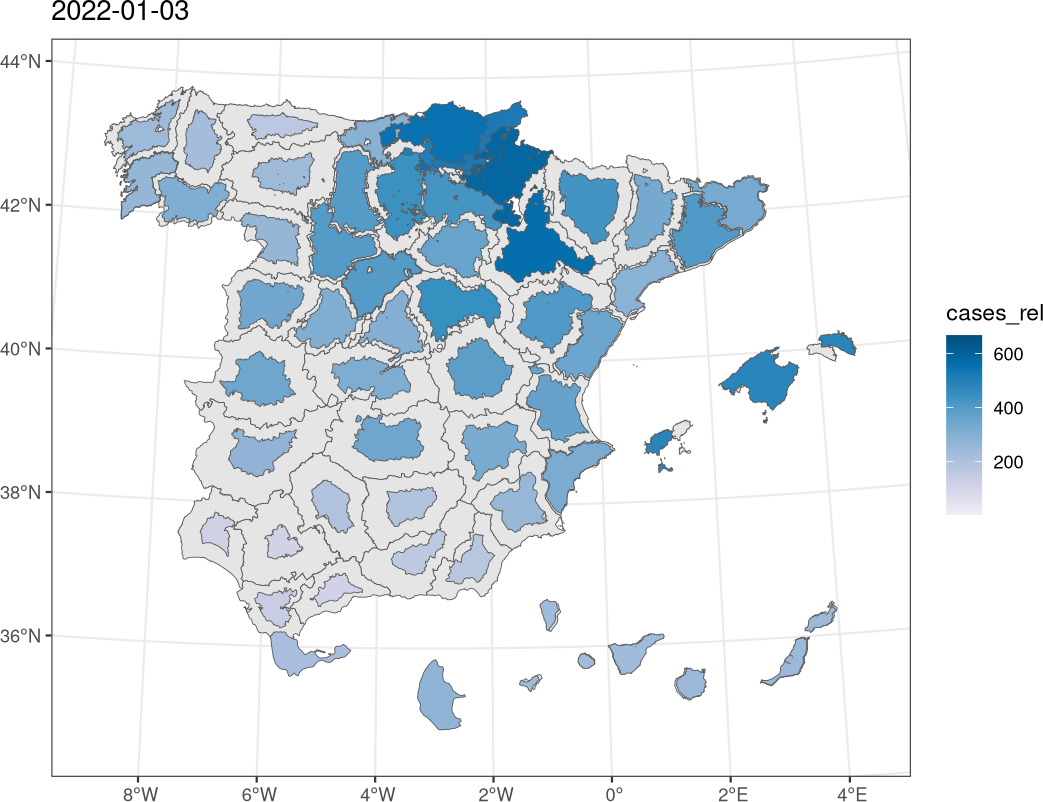
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. Sys.setlocale("LC_TIME", "C") ################################################################## ## Data and spatial information ################################################################## library("sf") library("cartogram") library("gganimate") covidSpain <- read.csv("data/SpatioTime/covid.csv", na.strings = NULL) covidSpain$PROV <- sprintf("%02d", covidSpain$PROV) covidSpain$day <- as.Date(covidSpain$day) covidSpain <- subset(covidSpain, day < as.Date("2022-03-01") & day >= as.Date("2021-12-01")) ## Population of each province popSpain <- read.csv("data/SpatioTime/PopSpain.csv") popSpain$PROV <- substring(popSpain$Province, 1, 2) popSpain2021 <- subset(popSpain, Year == 2021, select = c(PROV, Population)) covidSpain <- merge(covidSpain, popSpain2021, by = "PROV") ## Number of cases per 100.000 population covidSpain$cases_rel <- with(covidSpain, num_cases/Population * 1e5) ggplot(covidSpain) + geom_raster(aes(x = day, y = PROV, fill = cases_rel)) + scale_fill_distiller(palette = "PuBu", direction = 1) + theme_bw() sfProv <- st_read("data/Spatial/spain_provinces_2.shp", crs = 25830, stringsAsFactors = TRUE) ## Merge data with the polygons sfCovid <- merge(sfProv, covidSpain, by = "PROV") ggplot(subset(sfCovid, day >= as.Date("2022-01-01") & day <= as.Date("2022-01-09"))) + geom_sf(aes(fill = cases_rel)) + scale_fill_distiller(palette = "PuBu", direction = 1) + theme_bw() + facet_wrap(~ day, nrow = 3) ggCovid <- ggplot(sfCovid) + geom_sf(aes(fill = cases_rel)) + scale_fill_distiller(palette = "PuBu", direction = 1) + theme_bw() + ggtitle("{format(frame_time, format = '%Y-%m-%d')}") + transition_time(time = day) animate(ggCovid, height = 1080, width = 1080, res = 150, units = "px") fday <- as.Date("2022-01-01") lday <- as.Date("2022-02-28") days <- seq(fday, lday, by = "day") cartCOVIDList <- lapply(days, function(d) { x <- subset(sfCovid, day == d) cartogram_ncont(x, weight = "cases_rel") }) cartCOVID <- do.call(rbind, cartCOVIDList) ggplot(subset(cartCOVID, day >= as.Date("2022-01-01") & day <= as.Date("2022-01-09"))) + geom_sf(data = sfProv) + geom_sf(aes(fill = cases_rel)) + scale_fill_distiller(palette = "PuBu", direction = 1) + theme_bw() + facet_wrap(~ day, nrow = 3) ggCartCOVID <- ggplot(cartCOVID) + geom_sf(data = sfProv) + geom_sf(aes(fill = cases_rel, group = day)) + scale_fill_distiller(palette = "PuBu", direction = 1) + ggtitle("{format(frame_time)}") + transition_time(day) + theme_bw() animate(ggCartCOVID, height = 1080, width = 1080, res = 150, units = "px")
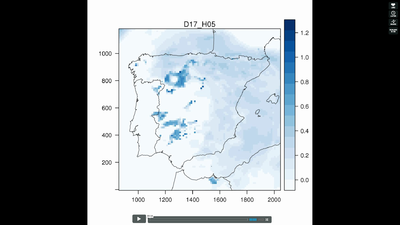
5.3. Spatiotemporal raster data
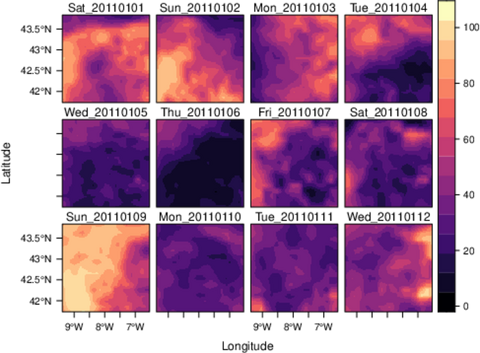
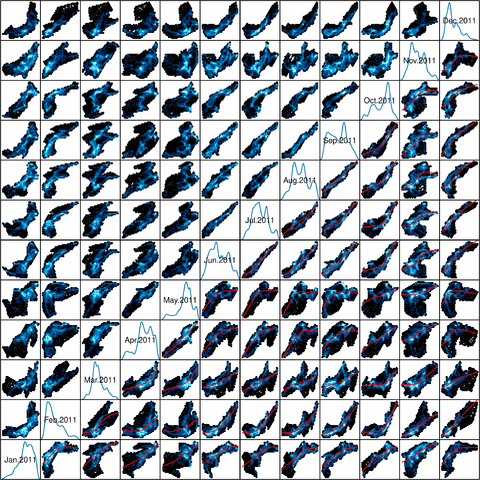
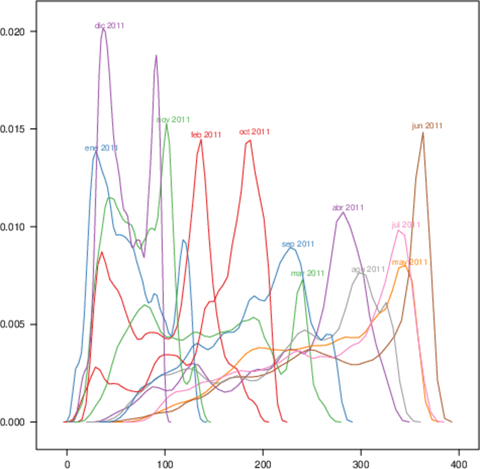
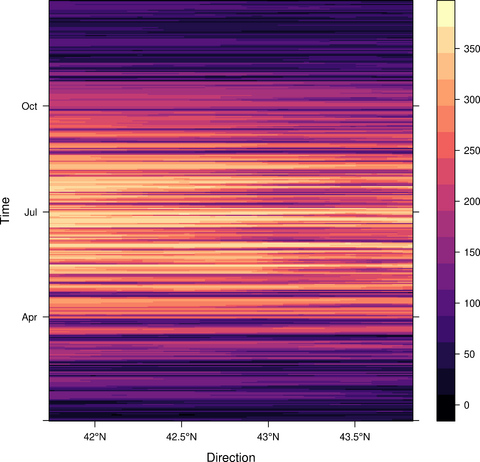
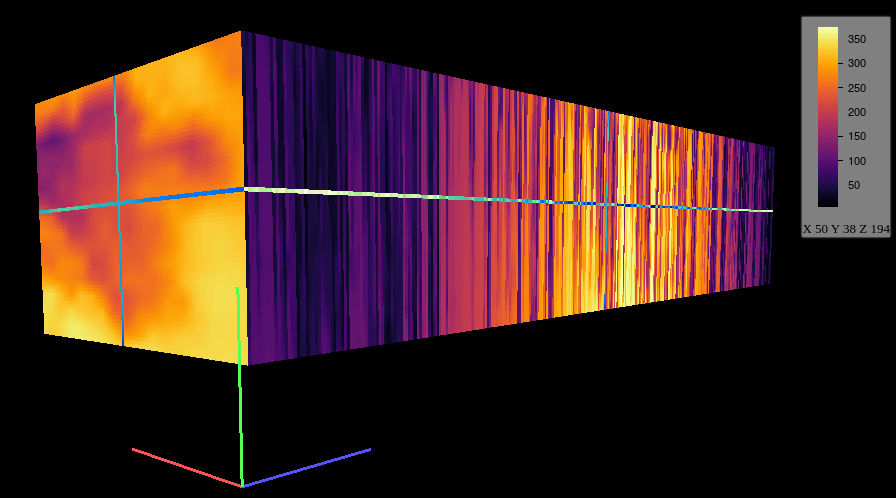
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. Sys.setlocale("LC_TIME", "C") library("raster") library("terra") library("zoo") library("RColorBrewer") library("rasterVis") SISdm <- brick("data/SpatioTime/SISgal") timeIndex <- seq(as.Date("2011-01-01"), by = "day", length = 365) SISdm <- setZ(SISdm, timeIndex) names(SISdm) <- format(timeIndex, "%a_%Y%m%d") SISdmt <- rast(SISdm) time(SISdmt) <- timeIndex ################################################################## ## Levelplot ################################################################## levelplot(SISdm, layers = 1:12, panel = panel.levelplot.raster) SISmm <- zApply(SISdm, by = as.yearmon, fun = 'mean') levelplot(SISmm, panel = panel.levelplot.raster) ################################################################## ## Exploratory graphics ################################################################## histogram(SISdm, FUN = as.yearmon) bwplot(SISdm, FUN = as.yearmon) splom(SISmm, xlab = '', plot.loess = TRUE) ################################################################## ## Space-time and time series plots ################################################################## hovmoller(SISdm) xyplot(SISdm, auto.key = list(space = 'right')) horizonplot(SISdm, digits = 1, col.regions = rev(brewer.pal(n = 6, 'PuOr')), xlab = '', ylab = 'Latitude') library("cubeview") ## cubeview has problems if the Raster* ## is not stored in memory SISdm <- readAll(SISdm) cubeview(SISdm) ################################################################## ## Data ################################################################## library("raster") library("rasterVis") cft <- brick("data/SpatioTime/cft_20130417_0000.nc") ## set projection projLCC2d <- "+proj=lcc +lon_0=-14.1 +lat_0=34.823 +lat_1=43 +lat_2=43 +x_0=536402.3 +y_0=-18558.61 +units=km +ellps=WGS84" projection(cft) <- projLCC2d ##set time index timeIndex <- seq(as.POSIXct("2013-04-17 01:00:00", tz = "UTC"), length = 96, by = "hour") cft <- setZ(cft, timeIndex) names(cft) <- format(timeIndex, "D%d_H%H") ################################################################## ## Spatial context: administrative boundaries ################################################################## library("rnaturalearth") library("sf") library("sp") world <- ne_countries(scale = "medium") ## Project the extent of the cft raster to longitude-latitude, because ## rnaturalearth works with it. cftLL <- projectExtent(cft, crs(world)) ## Crop... boundaries <- st_crop(world, cftLL) ## ... and project to the projection of the cft object boundaries <- st_transform(boundaries, crs(cft)) ## Finally, convert to a Spatial* object boundaries <- as(boundaries, "Spatial") ################################################################## ## Producing frames and movie ################################################################## library("RColorBrewer") library("latticeExtra") cloudTheme <- rasterTheme(region = brewer.pal(n = 9, 'Blues')) tmp <- tempdir() trellis.device(png, file = paste0(tmp, "/Rplot%02d.png"), res = 300, width = 1500, height = 1500) levelplot(cft, layout = c(1, 1), par.settings = cloudTheme, scales=list(draw=FALSE)) + layer(sp.lines(boundaries, lwd = 0.6)) dev.off() old <- setwd(tmp) ## Create a movie with ffmpeg ... system2("ffmpeg", c("-r 6", ## with 6 frames per second "-i Rplot%02d.png", ## using the previous files "-b:v 300k", ## with a bitrate of 300kbs "output.mp4") ) file.remove(dir(pattern = "Rplot")) file.copy("output.mp4", paste0(old, "/figs/SpatioTime/cft.mp4"), overwrite = TRUE) setwd(old) ################################################################## ## Static image ################################################################## levelplot(cft, layers = 25:48, ## Layers to display (second day) layout = c(6, 4), ## Layout of 6 columns and 4 rows par.settings = cloudTheme, scales=list(draw=FALSE), names.attr = paste0(sprintf("%02d", 1:24), "h"), panel = panel.levelplot.raster) + layer(sp.lines(boundaries, lwd = 0.6)) library("rgl") clear3d() pal <- colorRampPalette(brewer.pal(n = 9, "Blues")) N <- nlayers(cft) ids <- lapply(seq_len(N), FUN = function(i) plot3D(cft[[i]], maxpixels = 1e3, col = pal, adjust = FALSE, ## Disable automatic scaling of xy axes. zfac = 200)) ## Common z scale for all graphics library("manipulateWidget") rglwidget() %>% playwidget(start = 0, stop = N, subsetControl(1, subsets = ids))
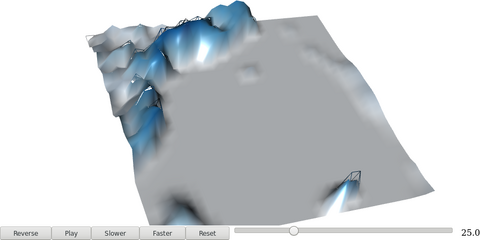
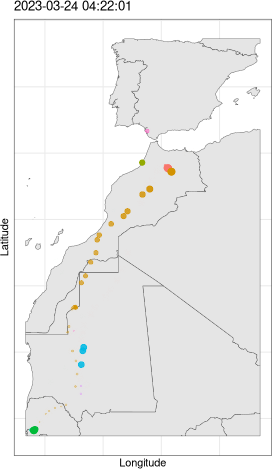
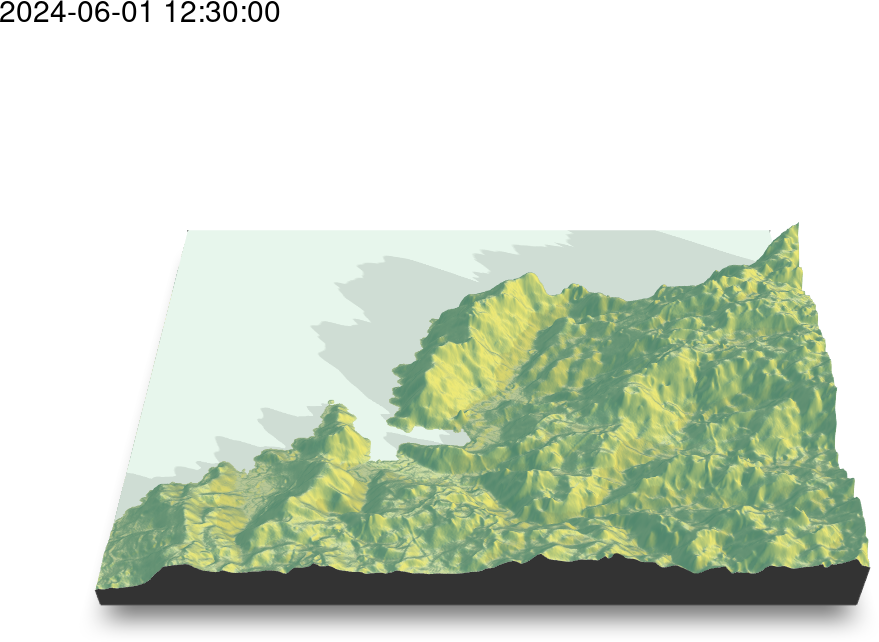
5.4. Animation
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. Sys.setlocale("LC_TIME", 'C') ################################################################## ## Time trajectory ################################################################## library("sf") library("move2") library("units") library("rnaturalearth") library("ggplot2") library("gganimate") ## Movebank data birds0 <- movebank_download_study(2398637362, "license-md5"="74263192947ce529c335a0ae72d7ead7") sf_use_s2(FALSE) ## Needed for st_crop to work ## Natural Earth boundaries boundaries <- ne_countries(scale = "large") boundaries <- st_crop(boundaries, birds0) ## Filter the data: speed higher than 2 m/s; remove year 2022 data. birds <- subset(birds0, ground_speed > set_units(2L, "m/s") & timestamp >= as.POSIXct("2023-01-01")) ## Add a column with month values birds$month <- as.numeric(format(mt_time(birds), "%m")) ggplot() + geom_sf(data = boundaries) + geom_sf(data = birds, aes(color = individual_local_identifier), alpha = 0.1) + guides(colour = guide_legend(override.aes = list(alpha = 1))) + theme_linedraw() + facet_wrap(~ month, nrow = 2) birds$speed <- cut(birds$ground_speed, breaks = c(2, 5, 10, 15, 35)) ggplot() + coord_polar(start = 0) + geom_histogram(data = birds, aes(x = set_units(heading, "degrees"), fill = speed), breaks = set_units(seq(0, 360, by = 10L), "degrees"), position = position_stack(reverse = TRUE)) + scale_x_units(name = NULL, limits = set_units(c(0L, 360), "degrees"), breaks = (0:4) * 90L) + ylab("") + facet_wrap(~ month, nrow = 2) + scale_fill_ordinal("Speed") + theme_linedraw() birdsMarch <- subset(birds, month == 3) p <- ggplot() + geom_sf(data = boundaries) + geom_sf(data = birdsMarch, aes(colour = individual_local_identifier), size = 3) + theme_bw() + xlab("Longitude") + ylab("Latitude") + ggtitle("{format(frame_time, format = '%Y-%m-%d %H:%M:%S')}") + transition_time(timestamp) + shadow_wake(0.8) animate(p, nframes = 300) ################################################################## ## Fly-by animation ################################################################## library("rgl") library("magick") ## needed to import the texture ## Opens the OpenGL device with a black background open3d() bg3d("black") ## XYZ coordinates of a sphere lat <- seq(-90, 90, len = 100) * pi/180 long <- seq(-180, 180, len = 100) * pi/180 r <- 6378.1 # radius of Earth in km x <- outer(long, lat, FUN = function(x, y) r * cos(y) * cos(x)) y <- outer(long, lat, FUN = function(x, y) r * cos(y) * sin(x)) z <- outer(long, lat, FUN = function(x, y) r * sin(y)) ## Read, scale, and convert the image nightLightsJPG <- image_read("https://eoimages.gsfc.nasa.gov/images/imagerecords/79000/79765/dnb_land_ocean_ice.2012.13500x6750.jpg") nightLightsJPG <- image_scale(nightLightsJPG, "8192") ## surface3d reads files up to 8192x8192 nightLights <- image_write(nightLightsJPG, tempfile(), format = "png") ## Only the png format is supported ## Display the sphere with the image superimposed surface3d(-x, -z, y, texture = nightLights, specular = "black", col = "white") rglwidget() cities <- rbind(c("Madrid", "Spain"), c("Tokyo", "Japan"), c("Sidney", "Australia"), c("Sao Paulo", "Brazil"), c("New York", "USA")) cities <- as.data.frame(cities) names(cities) <- c("city", "country") library("osmdata") geocode <- function(x) { place <- paste(x, collapse = ", ") bb <- getbb(place) center <- c(sum(bb[1, ])/2, sum(bb[2, ]/2)) center } points <- apply(cities, 1, geocode) points <- t(points) colnames(points) <- c("lon", "lat") cities <- cbind(cities, points) library("geosphere") ## When arriving or departing include a progressive zoom with 100 ## frames zoomIn <- seq(.3, .1, length = 100) zoomOut <- seq(.1, .3, length = 100) ## First point of the route route <- data.frame(lon = cities[1, "lon"], lat = points[1, "lat"], zoom = zoomIn, name = cities[1, "city"], action = "arrive") ## This loop visits each location included in the "points" set ## generating the route. for (i in 1:(nrow(cities) - 1)) { p1 <- cities[i,] p2 <- cities[i + 1,] ## Initial location departure <- data.frame(lon = p1$lon, lat = p1$lat, zoom = zoomOut, name = p1$city, action = "depart") ## Travel between two points: Compute 100 points between the ## initial and the final locations. routePart <- gcIntermediate(p1[, c("lon", "lat")], p2[, c("lon", "lat")], n = 100) routePart <- data.frame(routePart) routePart$zoom <- 0.3 routePart$name <- "" routePart$action <- "travel" ## Final location arrival <- data.frame(lon = p2$lon, lat = p2$lat, zoom = zoomIn, name = p2$city, action = "arrive") ## Complete route: initial, intermediate, and final locations. routePart <- rbind(departure, routePart, arrival) route <- rbind(route, routePart) } ## Close the travel route <- rbind(route, data.frame(lon = cities[i + 1, "lon"], lat = cities[i + 1, "lat"], zoom = zoomOut, name = cities[i+1, "city"], action = "depart")) summary(route) ## Function to move the viewpoint in the RGL scene according to the ## information included in the route (position and zoom). travel <- function(tt) { point <- route[tt,] view3d(theta = -90 + point$lon, phi = point$lat, zoom = point$zoom) par3d() } ## Example of usage of travel ## Frame no.1200 travel(1200) rgl.snapshot("figs/SpatioTime/rgl_travel1200.png") movie3d(travel, duration = nrow(route), startTime = 1, fps = 1, type = "mp4", clean = FALSE, webshot = FALSE) ################################################################## ## Hill shading animation ################################################################## library("rayshader") library("parallel") library("suntools") library("raster") library("gifski") demCedeira <- raster('data/Spatial/demCedeira') DEM <- raster_to_matrix(demCedeira) water <- detect_water(DEM) lonlat <- matrix(c((xmax(demCedeira) + xmin(demCedeira))/2, (ymax(demCedeira) + ymin(demCedeira))/2), nrow = 1) tt <- seq(as.POSIXct("2024-06-01 07:00:00", tz = "Europe/Madrid"), as.POSIXct("2024-06-01 21:00:00", tz = "Europe/Madrid"), by = "15 min") sun <- lapply(tt, function(x) solarpos(lonlat, x)) ncores <- detectCores() hillshades <- mclapply(sun, function(ang) { DEM %>% sphere_shade(texture = "imhof1", sunangle = ang[1]) %>% add_water(water, color = "imhof1") %>% add_shadow(ray_shade(DEM, sunangle = ang[1], sunaltitude = ang[2]), 0.75) }, mc.cores = ncores) old <- setwd(tempdir()) idx <- seq_along(hillshades) for(i in idx) { plot_3d(heightmap = DEM, hillshade = hillshades[[i]], zscale = 5, fov = 45, theta = 0, zoom = 0.75, phi = 45, windowsize = c(1000, 800)) render_snapshot(filename = paste0(i, ".png"), title_text = tt[i]) } gifski(png_files = paste0(idx, ".png"), gif_file = "cedeira.gif", delay = 1/5) setwd(old)